YOU ARE LEARNING:
Cutting DNA Out
Cutting DNA Out
Genetic engineering uses restriction enzymes to cut sections of DNA (genes) from human and other animal chromosomes, and ligase enzymes to join them to the DNA in another organism.
We use genetically engineered bacteria to produce medications, for example insulin.
The bacteria are genetically modified to carry the necessary insulin-producing gene from a human chromosome, but what piece of DNA from a bacteria do we insert the human gene into?
A) Bacterial plasmid B) Bacterial chromosome

All DNA, from all organisms has the same structure. It is made from a sequence of chemicals called nucleotides. Each one carries a base which acts as the "letters" of the code. What are the letters used for these 4 bases?

These bases pair up along the two strands of the DNA, so that each base only matches with one other base, but which ones match together?
If the sequence on one strand is: AAGCTT, what is the sequence on the opposite strand?

Remember that A pairs with T and G pairs with C.
So A is always opposite T and C is always opposite G.
To move a gene from a human chromosome to the plasmid of a bacteria, we have to make cuts in both the human and the bacterial DNA.
The cuts must happen at specific points along the DNA chain.
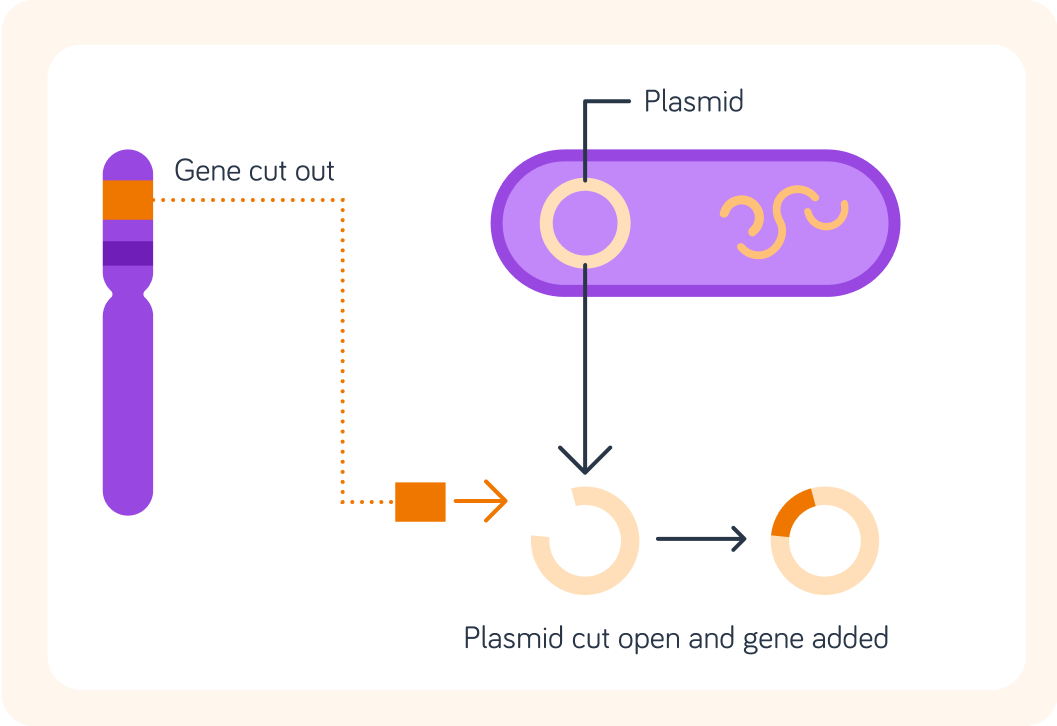
To do this we use a chemical called a restriction enzyme. Where would we need to make the cuts in the human DNA?
A) On either side of the chromosome B) On either side of the gene

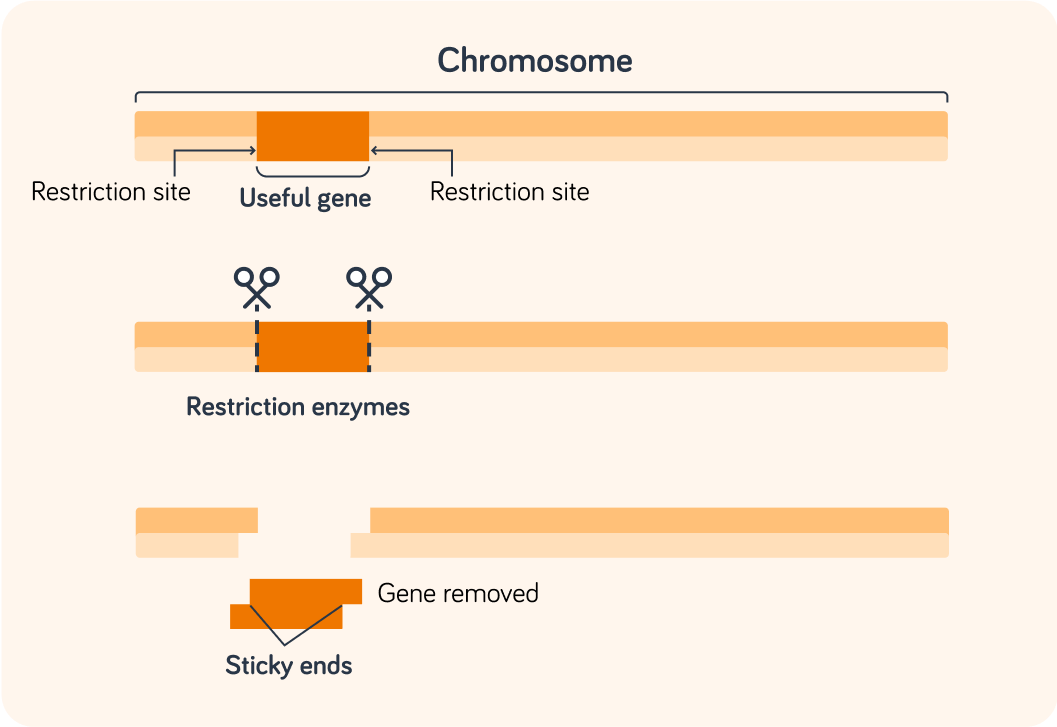
The useful gene is cut from the chromosome by two different restriction enzymes.
Each one targets one end of the gene.

Restriction enzymes target a specific sequence of bases on the DNA double strand. Why do you think that is important?

The restriction enzyme, HindIII, targets the base sequences shown in the diagram.
It will only cut this particular section and always in the way shown here.
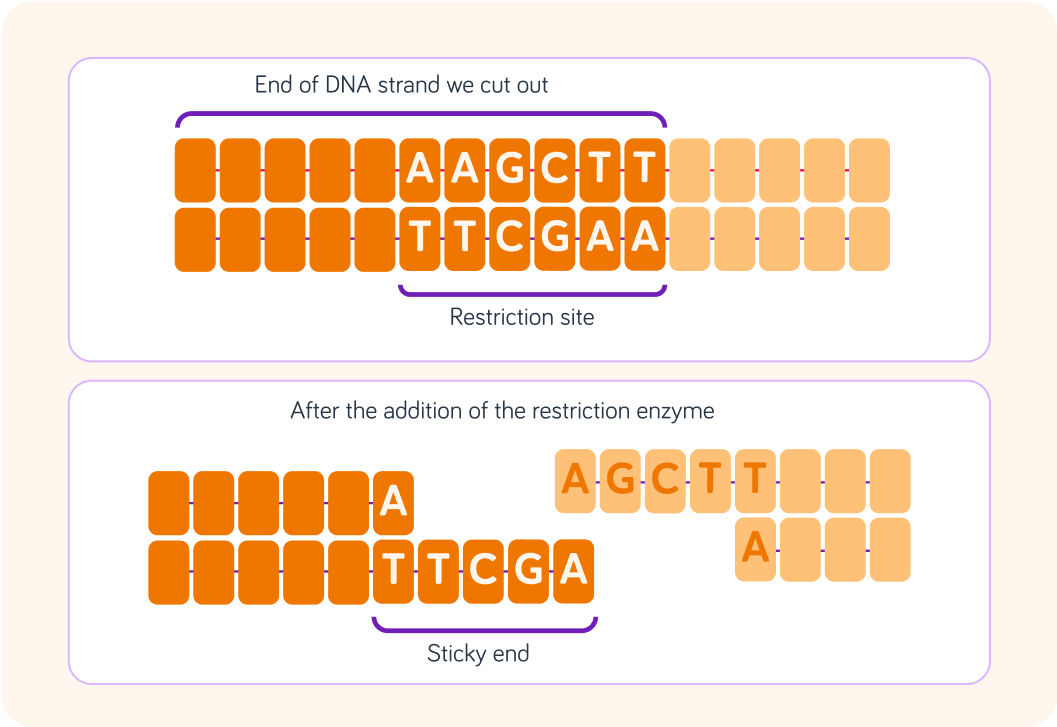
This leaves the unevenly cut ends where one is longer than the other.
These uneven end sequences are known as "sticky ends".

Other restriction enzymes recognise other base sequences like that in bold below and create a clean cut with a blunt end, for example...
CTAGG | CCTGA GATCC | GGACG

Now, we need to stick the gene into the plasmid.
We do this using a ligase enzyme.
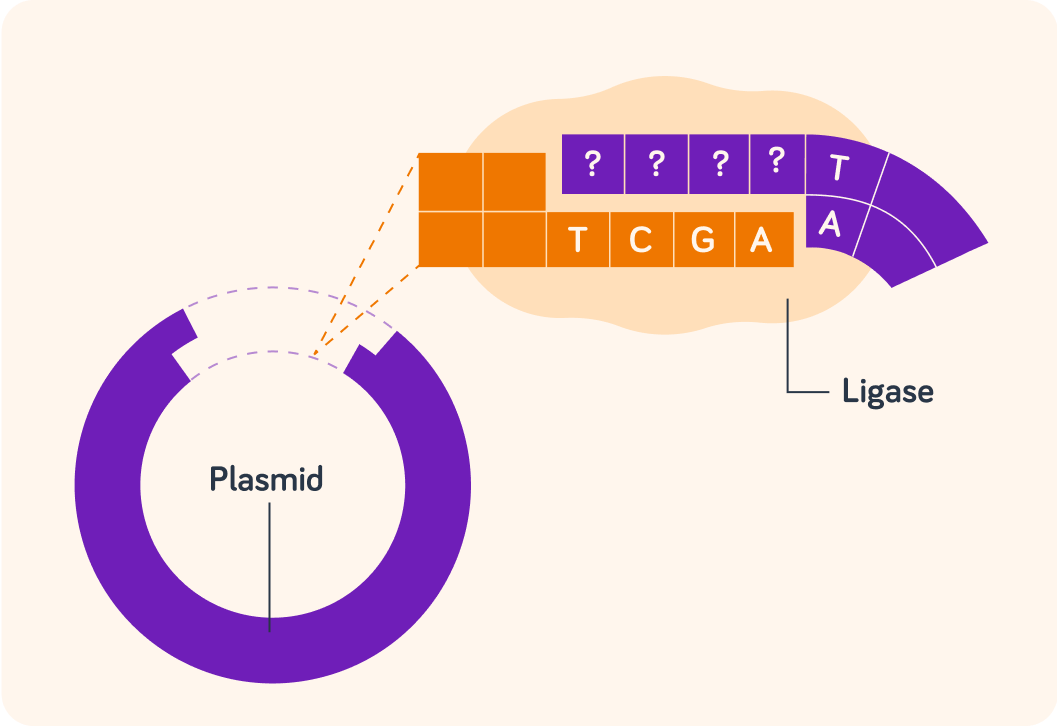
For this to work, the DNA sequence on the plasmid must match that on the cut gene. What must the top sequence of bases be on the sticky end of the plasmid for this gene to be attached successfully?


Restriction enzymes are used by bacteria to remove unwanted DNA that might attack them. For example, EcoRI enzymes comes form the bacteria E. coli. We get our restriction enzymes for genetic engineering from cultivating bacteria that have this ability.
